Tutorial: A Simple Vaccine Intervention
Day 4 | Network Modeling for Epidemics
EpiModel built-in network models also include parameters for a simple intervention that affects the probability of infection given contact between a susceptible and an infected person. This could be, for example, a vaccine that reduces the susceptibility or a condom provision program. This mini-tutorial will show you how to implement such an intervention. This is only meant to be used in the next lab. Research-level models would most likely implement more complex interventions with more parameters or greater demogrpahic structure; we’ll learn how to do that later today and tomorrow.
Interventions in this simple built-in approach have three salient features:
- They apply to everyone in the population. There is no heterogeneity in who gets the intervention.
- The interventions have an efficacy,
inter.eff, where the invention results in a relative reduction in the probability of infection specified ininf.probby1-inter.eff. - Interventions start at a defined time step,
inter.start.
To start, we estimate a very simple temporal ERGM.
nw <- network_initialize(n = 100, directed = FALSE)
formation <- ~edges
target.stats <- 50
coef.diss <- dissolution_coefs(dissolution = ~offset(edges), duration = 20)
est <- netest(nw, formation, target.stats, coef.diss, verbose = FALSE)This model simulates an SI disease in a closed population in which the intervention (e.g., a vaccine) has a very strong efficacy for the entire population. The intervention starts at week 25.
param <- param.net(inf.prob = 0.5, inter.eff = 0.96, inter.start = 25)
init <- init.net(i.num = 5)
control <- control.net(type = "SI", nsteps = 100, nsims = 10, ncores = 5)
sim <- netsim(est, param, init, control)
plot(sim)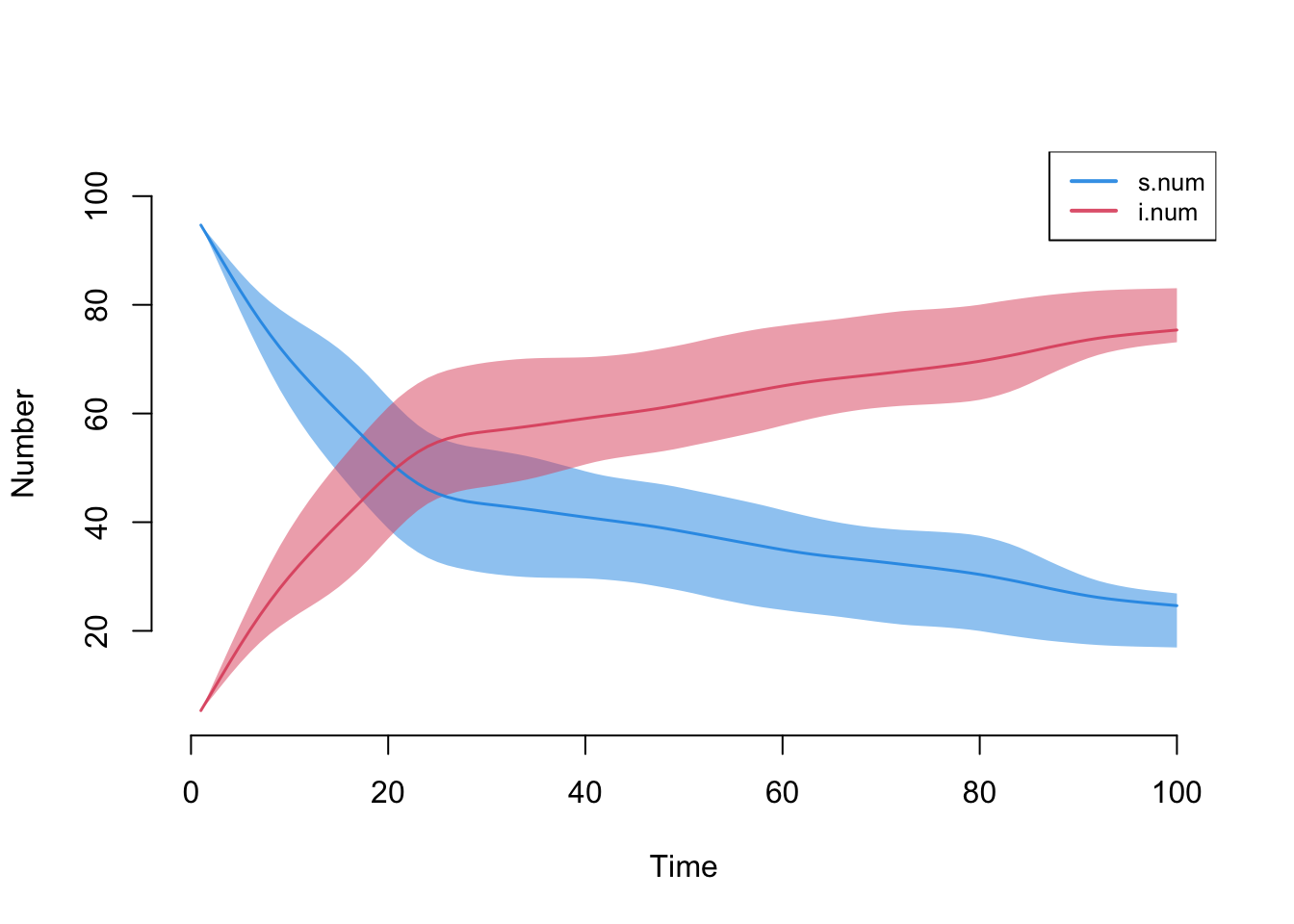
This model simulates an SIS disease in which the intervention is slightly less effective (e.g., functional effectiveness of condoms) that starts at week 100.
param <- param.net(inf.prob = 0.5, inter.eff = 0.8, inter.start = 100,
rec.rate = 0.07)
init <- init.net(i.num = 10)
control <- control.net(type = "SIS", nsteps = 250, nsims = 10, ncores = 5)
sim <- netsim(est, param, init, control)
plot(sim)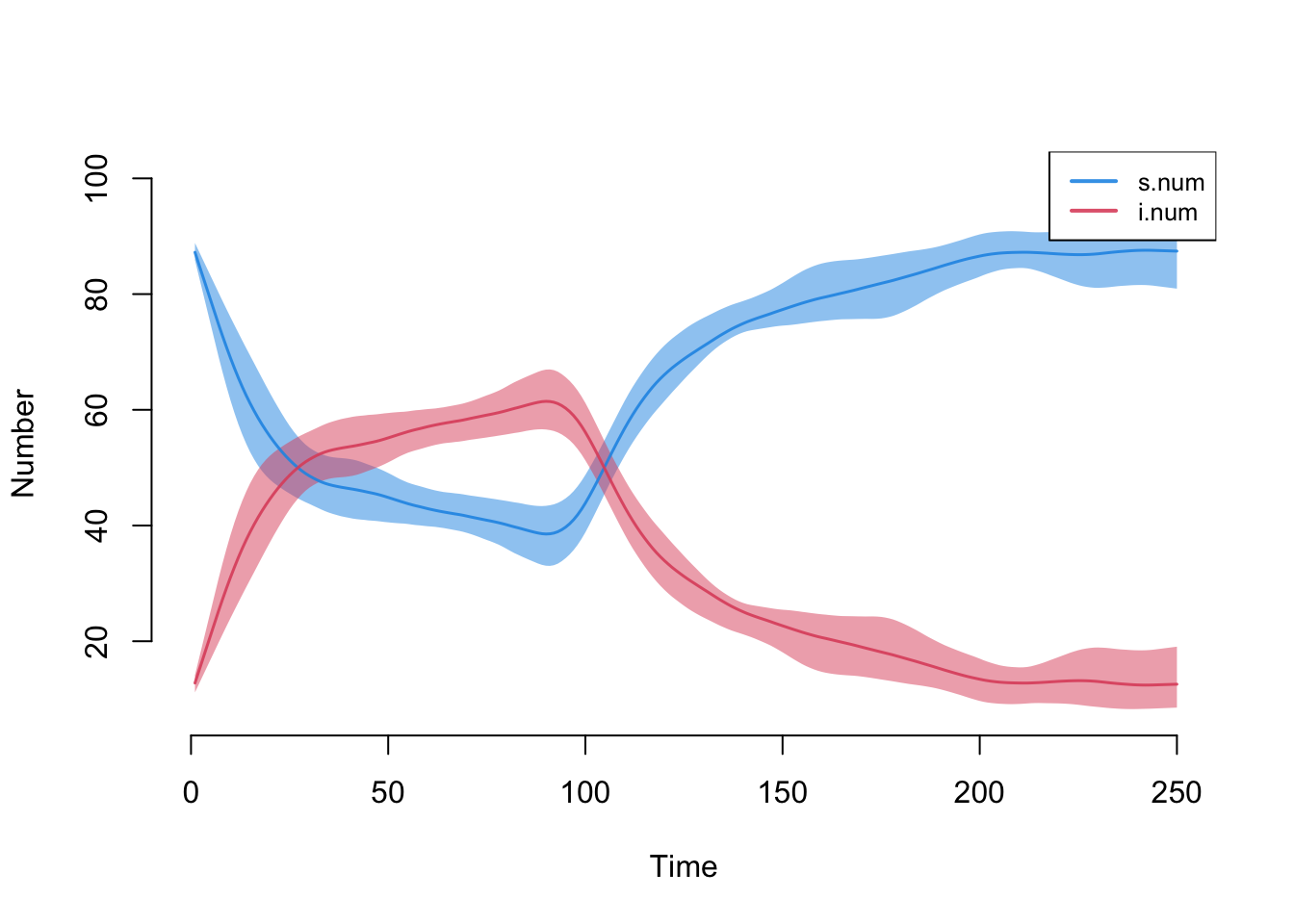
Last updated: 2022-07-07 with EpiModel v2.3.0